(Credit: Phys.org)
A new study has modeled a crucial first step in the self-assembly of cellular structures such as drug receptors and other protein complexes, and found that the flexibility of the structures has a dramatic impact on how fast two such structures join together.
The study, published this week in the journal Proceedings of the National Academy of Sciences, explored what happens when two water-repelling surfaces connect to build more complex structures. Using molecular simulations, the researchers illustrated the mechanism by which the process occurs and explored factors that favor self-assembly.
A surprise finding was the sensitivity with which the surfaces’ flexibility determined the rate at which the surfaces eventually came together, with more flexible surfaces favoring joining.
“Flexibility is like a knob that nature can tune to control the self-assembly of molecules,” says Pablo Debenedetti, senior author on the study and Princeton’s Dean for Research. Debenedetti is the Class of 1950 Professor in Engineering and Applied Science and a professor of chemical and biological engineering.
Researchers have long been interested in how biological structures can self-assemble according to physical laws. Tapping the secrets of self-assembly could, for example, lead to new methods of building nanomaterials for future electronic devices. Self-assembled protein complexes are the basis not only of drug receptors but also many other cellular structures, including ion channels that facilitate the transmission of signals in the brain.
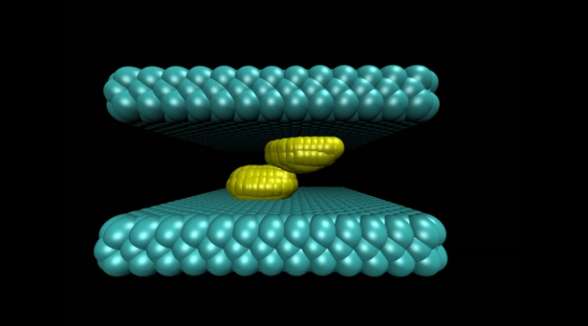
The study illustrated the process by which two water-repelling, or hydrophobic, structures come together. At the start of the simulation, the two surfaces were separated by a watery environment. Researchers knew from previous studies that these surfaces, due to their hydrophobic nature, will push water molecules away until only a very few water molecules remain in the gap. The evaporation of these last few molecules allows the two surfaces to snap together.
The new molecular simulation conducted at Princeton yielded a more detailed look at the mechanism behind this process. In the simulation, when the surfaces are sufficiently close to each other, their hydrophobic nature triggered fluctuations in the number of water molecules in the gap, causing the liquid water to evaporate and form bubbles on the surfaces. The bubbles grew as more water molecules evaporated. Eventually two bubbles on either surface connected to form a gap-spanning tube, which expanded and pushed away any remaining water until the two surfaces collided.
Biological surfaces, such as cellular membranes, are flexible, so the researchers explored how the surfaces’ flexibility affected the process. The researchers tuned the flexibility of the surfaces by varying the strength of the coupling between the surface atoms. The stronger the coupling, the less each atom can wiggle relative to its neighbors.
The researchers found that the speed at which the two surfaces snap together depended greatly on flexibility. Small changes in flexibility led to large changes in the rate at which the surfaces stuck together. For example, two very flexible surfaces adhered in just nanoseconds, whereas two inflexible surfaces fused incredibly slowly, on the order of seconds.
Another finding was that the last step in the process, where the vapor tube expands, was critical for ensuring that the surfaces came together. In simulations where the tube failed to expand, the surfaces never joined. Flexibility was key to ensuring that the tube expanded, the researchers found. Making the material more flexible lowered the barriers to evaporation and stabilized the vapor tube, increasing the chances that the tube would expand.
The molecular simulation provides a foundation for understanding how biological structures assemble and function, according to Elia Altabet, a graduate student in Debenedetti’s group, and first author on the study. “A deeper understanding of the formation and function of protein assemblies such as drug receptors and ion channels could inform the design of new drugs to treat diseases,” he says.


