For the first time, Massachusetts Institute of Technology (MIT) engineers have designed sensors that can detect single protein molecules as they are secreted by cells or even a single cell.
These sensors, which consist of chemically modified carbon nanotubes, could help scientists with any application that requires detecting very small amounts of protein, such as tracking viral infection, monitoring cells’ manufacturing of useful proteins, or revealing food contamination, the researchers say.
“We hope to use sensor arrays like this to look for the ‘needle in a haystack,'” says Michael Strano, the Carbon P. Dubbs Professor of Chemical Engineering at MIT. “These arrays represent the most sensitive molecular sensing platforms that we have available to us technologically. You can functionalize them so you can see the stochastic fluctuations of single molecules binding to them.”
Strano is the senior author of a Jan. 23 Nature Nanotechnology paper describing the new sensors. The paper’s lead author is Markita Landry, a former MIT postdoc who is now an assistant professor at the University of California at Berkeley.
Other MIT authors are research scientist Hiroki Ando, former graduate student Allen Chen, postdocs Jicong Cao and Juyao Dong, and associate professor of electrical engineering and computer science Timothy Lu. Vishal Kottadiel of Harvard University and Linda Chio and Darwin Yang of the University of California at Berkeley are also authors.
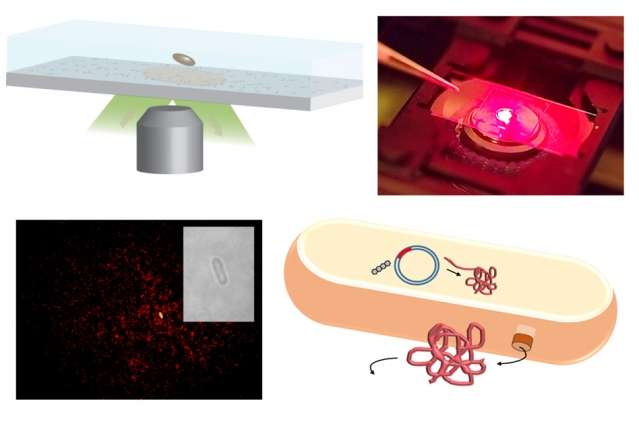
MIT chemical engineers have developed arrays of carbon nanotube sensors that can detect single protein molecules as they are secreted from cells. (Credit: Massachusetts Institute of Technology)
No detection limit
Strano’s lab has previously developed sensors that can detect many types of molecules, all based on modifications of carbon nanotubes—hollow, nanometer-thick cylinders made of carbon that naturally fluoresce when exposed to laser light. To turn the nanotubes into sensors, Strano’s lab coats them with DNA, proteins, or other molecules that can bind to a specific target. When the target is bound, the nanotubes’ fluorescence changes in a measurable way.
In this case, the researchers used chains of DNA called aptamers to coat the carbon nanotubes. Previous efforts to use DNA aptamers have been stymied because of the difficulty of getting the aptamer to stick to the nanotube while maintaining the configuration it needs to bind to its target.
Landry overcame this challenge by adding a “spacer” sequence between the section of the aptamer that attaches to the nanotube and the section that binds to the target, allowing each region the freedom to perform its own function. The researchers successfully demonstrated sensors for a signaling protein called RAP1 and a viral protein called HIV1 integrase, and they believe the approach should work for many other proteins.
To monitor protein production of single cells, the researchers set up an array of the sensors on a microscope slide. When a single bacterial, human, or yeast cell is placed on the array, the sensors can detect whenever the cell secretes a molecule of the target protein.
“Nanosensor arrays like this have no detection limit,” Strano says. “They can see down to single molecules.”
However, there is a tradeoff—the fewer molecules there are, the longer it takes to sense them. As the molecule becomes more scarce, detection can take an infinite amount of time, Strano says.
“The new study by Strano and co-workers proposes an exciting new approach to detect proteins down to the single molecule level,” says Robert Hurt, a professor of engineering at Brown University who was not involved in the research. “The work pushes the forefront in single-protein detection and may allow researchers to see important, real-time molecular events at the single-cell level, such as protein release during cell division.”
Useful tools
The sensor arrays could be useful for many different applications, the researchers say.
“This platform will open a new path to detect trace amounts of proteins secreted by microorganisms,” Dong says. “It will advance biological research [on] the generation of signal molecules, as well as the biopharmaceutical industry’s [efforts to monitor] microorganism health and product quality.”
In the pharmaceutical realm, these sensors could be used to test cells engineered to help treat disease. Many researchers are now working on an approach where doctors would remove a patient’s own cells, engineer them to express a therapeutic protein, and place them back in the patient.
“We think these nanosensor arrays are going to be useful tools for measuring these precious cells and making sure that they’re performing the way that you want them to,” Strano says.
He says researchers could also use the arrays to study viral infection, neurotransmitter function, and a phenomenon called quorum sensing, which allows bacteria to communicate with each other to coordinate their gene expression.


