A low-cost technology may make it possible to read long sequences of DNA far more quickly than current techniques.
A low-cost technology may make it possible to read long sequences of DNA far more quickly than current techniques.
The research advances a technology, called nanopore DNA sequencing. If perfected it could someday be used to create handheld devices capable of quickly identifying DNA sequences from tissue samples and the environment, the University of Washington researchers who developed and tested the approach said.
“One reason why people are so excited about nanopore DNA sequencing is that the technology could possibly be used to create ‘tricorder’-like devices for detecting pathogens or diagnosing genetic disorders rapidly and on-the-spot,” said Andrew Laszlo, lead author and a graduate student in the laboratory of Jen Gundlach, a UW professor of physics who led the project.
The paper “Decoding long nanopore sequencing reads of natural DNA” describes the new technique. It appears June 25 in the advanced online edition of the journal Nature Biotechnology.
Most of the current gene sequencing technologies require working with short snippets of DNA, typically 50 to 100 nucleotides long. These must be processed by large sequencers in a laboratory. The cumbersome process can take days to weeks to complete.
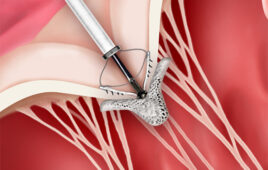
Nanopore technology takes advantage of the small, tunnel-like structures found in bacterial membranes. In nature, such pores allow bacteria to control the flow of nutrients across their membranes.
UW researcher used the nanopore Mycobacterium smegmatis porin A (MspA). This bacterial pore has been genetically altered so that the narrowest part of the channel has a diameter of about a nanometer, or 1 billionth of a meter. This is large enough for a single strand of DNA to pass through. The modified nanopore is then inserted into a membrane separating two salt solutions to create a channel connecting the two solutions.
To read a sequence of DNA with this system, a small voltage is applied across the membrane to make the ions of the salt solution flow through the nanopore. The ion flow creates a measurable current. If a strand of DNA is added to the solution on one side of the membrane and then enters a pore, the bulky DNA molecules will impede the flow of the much smaller ion and thereby alter the current. How much the current changes depends on which nucleotides — the individual molecules adenine, guanine, cytosine and thymine that make up the DNA chain — are inside the pore. Detecting changes in current can reveal which nucleotides are passing through the nanopore’s channel at any given instant.
Since the technique was first proposed in the 1990s, researchers hoped that nanopore DNA sequencing would offer a cheap, fast alternative to current gene sequencing. But their attempts have been frustrated by several challenges. It is difficult to identify each nucleotide one-by-one as they pass through the nanopore. Instead, researchers have to work with changes in current associated with four nucleotides at a time. In addition, some nucleotides may be missed or read more than once. Consequently, current nanopore sequencing technology yields an imprecise readout of a DNA sequence.
The UW researchers describe how they bypassed these problems. The researchers first identified the electronic signatures of all the nucleotide combinations possible with the four nucleotides that make up DNA — a total of 256 combinations in all (4 x 4 x 4 x 4).
They then created computer algorithms to match the current changes generated when a segment of DNA passes through the pore with current changes expected from DNA sequences of known genes and genomes stored in a computer database. A match would show that the sequence of the DNA passing through the pore was identical or close to the DNA sequence stored in the database. The whole process would take minutes to a few hours, instead of weeks.
To test this approach, the researchers used their nanopore system to read the sequence of bacteriophage Phi X 174, a virus that infects bacteria and that is commonly used to evaluate new genome sequencing technologies. They found that the approach reliably read the bacteriophage’s DNA sequences and could read sequences as long as 4,500 nucleotides.
“This is the first time anyone has shown that nanopores can be used to generate interpretable signatures corresponding to very long DNA sequences from real-world genomes,” said co-author Jay Shendure, a UW associate professor of genome sciences whose lab develops applications of genome sequencing technologies. “It’s a major step forward.”
Because the technique relies on matching readings to databases of previously sequenced genes and genomes, it cannot yet be used to sequence a newly discovered gene or genome, the researchers said, but with some refinements, they added, it should be possible to improve performance in this area. To accelerate research on this new technology, the scientists are making their methods, data and computer algorithms fully available to all.
“Despite the remaining hurdles, our demonstration that a low-cost device can reliably read the sequences of naturally occurring DNA and can interpret DNA segments as long as 4,500 nucleotides in length represents a major advance in nanopore DNA sequencing,” Gundlach said.
This work was supported by the National Institutes of Health, National Human Genome Research Institutes $1,000 Genome Program Grants R01HG005115, R01HG006321 and R01HG006283 and a graduate research fellowship from the National Science Foundation DGE-0718124.