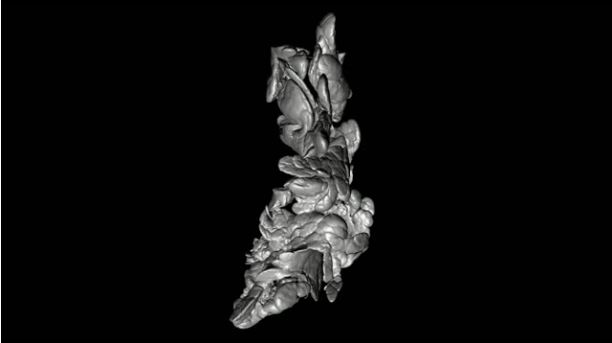
(Credit: Ulf Ahlgren)
Umeå researchers have created datasets that map the 3D distribution and volume of the insulin-producing cells in the pancreas. The wealth of visual and quantitative information may serve as powerful reference resource for diabetes researchers.
The Umeå University researchers are now publishing their datasets in Scientific Data, which is a Nature Research journal for scientifically valuable collections of research data with high reuse potential.
The hormone insulin — which is needed to regulate the blood sugar levels of the body — is produced by the pancreas and plays a key role in the development of diabetes. Insulin-producing cells are organised in the so-called Islets of Langerhans (or pancreatic islets), which are scattered by the thousands in the pancreas. In diabetes research, it is often important to study the quantity and distribution of insulin-producing cells. At present, such studies are generally based upon analyses of chosen cross-sections of pancreatic tissue. These in turn form the basis for attempting to gain an overall picture of the pancreas.
“However, such analyses only provide limited information and are often ridden with relatively large margins of error since the conclusions are based only on two-dimensional data,” says Ulf Ahlgren, professor in molecular medicine at Umeå University and in charge of the publications.
Ulf Ahlgren and his research colleagues at the Umeå Centre for Molecular Medicine (UCMM) have previously developed new methods to create three-dimensional images of the insulin cell distribution in intact pancreas based on so-called optical projection tomography (OPT). This technique in many ways bears resemblance to a medical CT scanner, but instead of x-rays it uses regular light.
“We believe that the current publication represents the most comprehensive anatomical and quantitative description of the insulin cell distribution in the pancreas. By making these datasets accessible to other researchers, the data will be available for use as a powerful tool for a great number of diabetes studies. Examples may include planning of stereological analyses, in the development of non-invasive imaging techniques or various types of computational modelling and statistical analyses,” says Ulf Ahlgren.
The datasets now published in Scientific Data consist of tomographic and 3D images. The datasets also include information on the individual volume of the Islets of Langerhans and their 3D coordinates and appearance throughout the entire pancreas in both healthy mice and obese mice (ob/ob), at different ages. The obese mice used in the study have a mutation that make them prone to develop obesity and diabetes.
The datasets highlight that islets differ in size and quantity within, and between, the various lobes of the pancreas. According to the research team, this emphasises that the pancreas should not be seen as a homogenous organ when experimental diabetes researchers study the insulin-producing Islets of Langerhans.
Visualising changes in the Islets of Langerhans
The datasets presented in Scientific Data form the basis of another recently published study in Scientific Reports. In that study, the researchers used the 3D data to identify changes in the Islets of Langerhans in the obese (ob/ob) mice. This animal model is often used to study initial metabolic changes that can lead to the development of type 2 diabetes. With the help of their refined techniques, the researchers could show that these mice to a great extent develop lesions in the Islets of Langerhans, manifesting as cyst-like structures. The study shows that these lesions are caused by internal bleeding as a consequence of an increased blood flow and instability of the blood vessels.
“Obese (ob/ob) mice have been described in thousands of publications. But the large prevalence of such internal islet lesions have never before been identified and visualised,” says Ulf Ahlgren.
The researchers now want to study if similar intra-islet lesions also form in other models of type 2 diabetes and in humans, and if these may contribute to the diabetic phenotype.




